Quickstart¶
[1]:
%load_ext autoreload
%autoreload 2
[2]:
from os import environ
GOOGLE_COLAB = True if "COLAB_GPU" in environ else False
if GOOGLE_COLAB:
!pip install git+https://github.com/undark-lab/swyft.git
[3]:
import numpy as np
import pylab as plt
import swyft
Setup the forward model¶
[4]:
device = "cpu"
n_training_samples = 10_000
n_parameters = 2
marginal_indices_1d, marginal_indices_2d = swyft.utils.get_corner_marginal_indices(n_parameters)
observation_key = "x"
n_weighted_samples = 10_000
[5]:
def model(v, sigma = 0.2):
x = v + np.random.randn(n_parameters) * sigma
return {observation_key: x}
v_o = np.zeros(n_parameters)
observation_o = model(v_o, sigma = 0.)
n_observation_features = observation_o[observation_key].shape[0]
observation_shapes = {key: value.shape for key, value in observation_o.items()}
[6]:
simulator = swyft.Simulator(
model,
n_parameters,
sim_shapes=observation_shapes
)
Setup the prior and storage¶
[7]:
low = -1 * np.ones(n_parameters)
high = 1 * np.ones(n_parameters)
prior = swyft.get_uniform_prior(low, high)
store = swyft.Store.memory_store(simulator)
store.add(n_training_samples, prior)
store.simulate()
Creating new store.
Store: Adding 9746 new samples to simulator store.
[8]:
dataset = swyft.Dataset(n_training_samples, prior, store)
Train a 1d marginal estimator¶
[9]:
network_1d = swyft.get_marginal_classifier(
observation_key=observation_key,
marginal_indices=marginal_indices_1d,
observation_shapes=observation_shapes,
n_parameters=n_parameters,
hidden_features=32,
num_blocks=2,
)
mre_1d = swyft.MarginalRatioEstimator(
marginal_indices=marginal_indices_1d,
network=network_1d,
device=device,
)
[10]:
mre_1d.train(dataset)
training: lr=5e-06, epoch=25, validation loss=1.6534
Create a simple violin plot and a row of histograms to view the 1d marginals¶
[11]:
n_rejection_samples = 5_000
posterior_1d = swyft.MarginalPosterior(mre_1d, prior)
samples_1d = posterior_1d.sample(n_rejection_samples, observation_o)
[12]:
_ = swyft.violin(samples_1d)
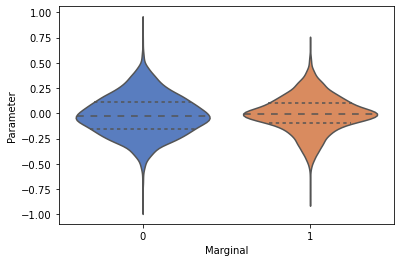
[13]:
_, _ = swyft.hist1d(samples_1d, kde=True)
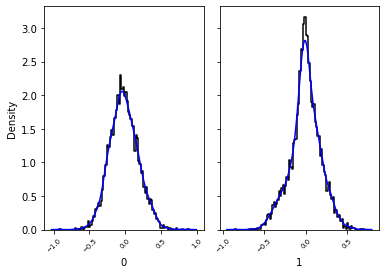
Train a 2d marginal estimator¶
[14]:
network_2d = swyft.get_marginal_classifier(
observation_key=observation_key,
marginal_indices=marginal_indices_2d,
observation_shapes=observation_shapes,
n_parameters=n_parameters,
hidden_features=32,
num_blocks=2,
)
mre_2d = swyft.MarginalRatioEstimator(
marginal_indices=marginal_indices_2d,
network=network_2d,
device=device,
)
[15]:
mre_2d.train(dataset)
training: lr=5e-07, epoch=25, validation loss=0.4701
Combine the two to create a corner plot¶
[16]:
posterior_1d = swyft.MarginalPosterior(mre_1d, prior)
weighted_samples_1d = posterior_1d.weighted_sample(n_weighted_samples, observation_o)
posterior_2d = swyft.MarginalPosterior(mre_2d, prior)
weighted_samples_2d = posterior_2d.weighted_sample(n_weighted_samples, observation_o)
[17]:
_, _ = swyft.corner(
weighted_samples_1d,
weighted_samples_2d,
kde=True,
truth=v_o,
labels=["x1", "x2"]
)
1it [00:04, 4.01s/it]
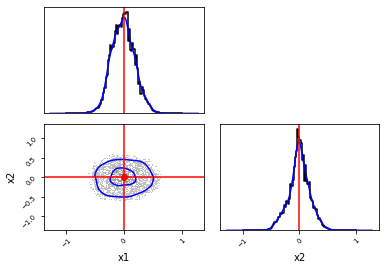
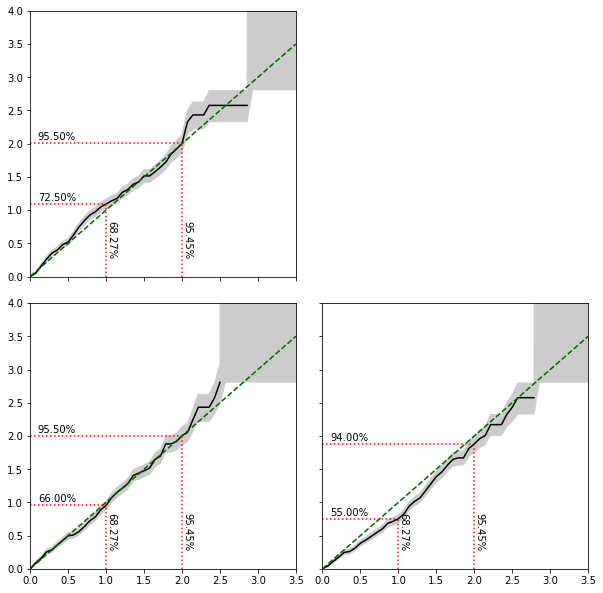
Test the expected coverage probability¶
[18]:
n_observations = 200
n_posterior_samples = 5_000
empirical_mass_1d, _ = posterior_1d.empirical_mass(n_observations, n_posterior_samples, dataset)
empirical_mass_2d, _ = posterior_2d.empirical_mass(n_observations, n_posterior_samples, dataset)
[19]:
_, _ = swyft.empirical_z_score_corner(empirical_mass_1d, empirical_mass_2d, figsize=(10,10))