This page was generated from notebooks/0E - Coverage tests.ipynb.
E- Coverage tests#
Authors: Noemi Anau Montel, James Alvey, Christoph Weniger
Last update: 27 April 2023
Purpose: Testing posteriors and inference results.
Key take-away messages: How can we generate pp and zz plot with swyft?
Code#
[ ]:
import numpy as np
import pylab as plt
import torch
import swyft
We again consider the same linear regression problem as above…
[ ]:
N = 10_000 # Number of samples
Nbins = 100
z = np.random.rand(N, 3)*2 - 1
y = np.linspace(-1, 1, Nbins)
m = np.ones_like(y)*z[:,:1] + y*z[:,1:2] + y**2*z[:,2:]
x = m + np.random.randn(N, Nbins)*0.2 #+ np.random.poisson(lam = 3/Nbins, size = (N, Nbins))*5
# We keep the first sample as observation, and use the rest for training
samples = swyft.Samples(x = x, z = z)
obs = swyft.Sample(samples[0])
…as well as an inference network with linear data summaries, …
[ ]:
class Network(swyft.SwyftModule):
def __init__(self):
super().__init__()
self.summarizer = torch.nn.Linear(Nbins, 3)
self.logratios = swyft.LogRatioEstimator_1dim(num_features = 1, num_params = 3, varnames = 'z')
def forward(self, A, B):
s = self.summarizer(A['x'])
s = s.unsqueeze(-1)
return self.logratios(s, B['z'])
…and train our inference network as usual.
[ ]:
trainer = swyft.SwyftTrainer(accelerator = 'gpu', devices=1, max_epochs = 3, precision = 64)
dm = swyft.SwyftDataModule(samples[1:-500], fractions = [0.8, 0.2, 0.0])
network = Network()
trainer.fit(network, dm)
INFO:pytorch_lightning.utilities.rank_zero:GPU available: True (cuda), used: True
INFO:pytorch_lightning.utilities.rank_zero:TPU available: False, using: 0 TPU cores
INFO:pytorch_lightning.utilities.rank_zero:IPU available: False, using: 0 IPUs
INFO:pytorch_lightning.utilities.rank_zero:HPU available: False, using: 0 HPUs
INFO:pytorch_lightning.accelerators.cuda:LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
INFO:pytorch_lightning.callbacks.model_summary:
| Name | Type | Params
------------------------------------------------------
0 | summarizer | Linear | 303
1 | logratios | LogRatioEstimator_1dim | 52.2 K
------------------------------------------------------
52.5 K Trainable params
0 Non-trainable params
52.5 K Total params
0.420 Total estimated model params size (MB)
INFO:pytorch_lightning.utilities.rank_zero:`Trainer.fit` stopped: `max_epochs=3` reached.
Like above, let’s look at the posteriors for a specific mock observation obs.
[ ]:
prior_samples = swyft.Samples(z = np.random.rand(10_000, 3)*2-1)
predictions = trainer.infer(network, obs, prior_samples)
fig, axes = plt.subplots(1, 3, figsize = (12, 4))
for i in range(3):
swyft.plot_1d(predictions, "z[%i]"%i, ax = axes[i]); axes[i].axvline(obs['z'][i], color='r'); axes[i].set_xlabel("z[%i]"%i)
plt.tight_layout();
INFO:pytorch_lightning.accelerators.cuda:LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
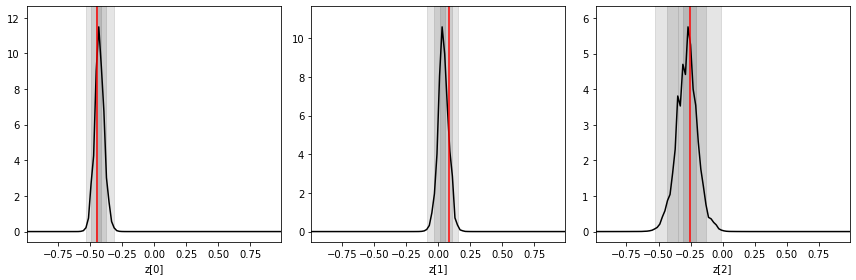
How do we know that these posteriors are “correct”? Let us now consider Bayesian coverage. To this end, we use the last 500 samples and estimate the Bayesian coverage (the fraction of samples where the \(1-\alpha\) credible region contains the true value – which should be \(1-\alpha\)).
[ ]:
coverage_samples = trainer.test_coverage(network, samples[-500:], prior_samples)
INFO:pytorch_lightning.accelerators.cuda:LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
WARNING: This estimates the mass of highest-likelihood intervals.
INFO:pytorch_lightning.accelerators.cuda:LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
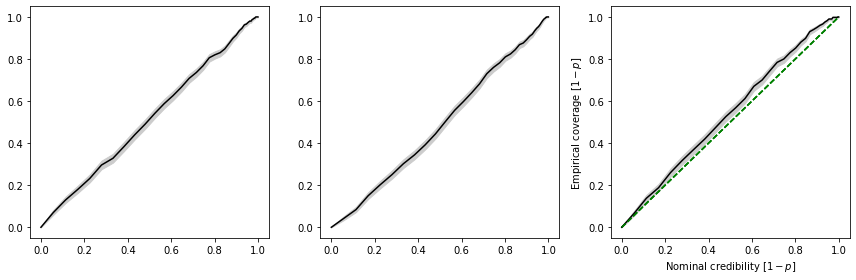
The coverage test restults can be visualized using swyft.plot_pp, which generates the standard pp-plot commonly seen in the literature. The disadvantage of this plot is that it squashes the particularly interesting high-credibility regions together.
[ ]:
fix, axes = plt.subplots(1, 3, figsize = (12, 4))
for i in range(3):
swyft.plot_pp(coverage_samples, "z[%i]"%i, ax = axes[i])
plt.tight_layout()
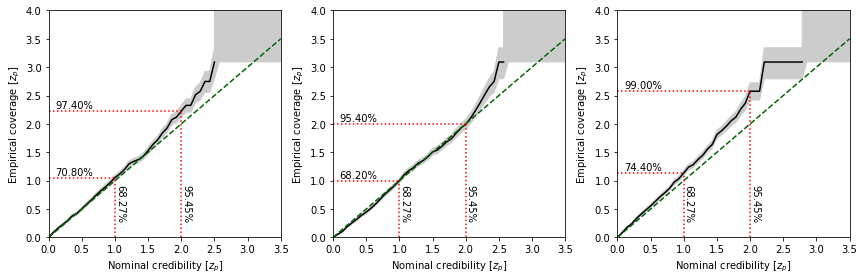
Instead, in Swyft we use something we decided to call zz-plots, swyft.plot_zz, which emphasises high credibility regions and visualizes the uncertainties of the coverage test which are associated with a finite sample size (in this example just 500).
[ ]:
fix, axes = plt.subplots(1, 3, figsize = (12, 4))
for i in range(3):
swyft.plot_zz(coverage_samples, "z[%i]"%i, ax = axes[i])
plt.tight_layout()
Exercises#
Check how coverage changes when reducing or increasing the number of epochs during training. An overtrained network will have overconfident coverage.
[ ]:
# Your results here
This page was generated from notebooks/0E - Coverage tests.ipynb.